The GithubMetrics package
Easy access to GithubMetrics via a gh wrapper
At work I manage a data science team, and the backbone to our work is an
on-premise Github server. This holds our research code, as well as
pan-study code (e.g. packages and libraries). To help keep on top of our
codebase, we use the Github API. To make it easier to manage this
codebase, I threw some of these functions into an R package called
GithubMetrics.
- Package: https://github.com/openpharma/GithubMetrics
- Docs: https://openpharma.github.io/GithubMetrics/
The aim of this package is to provide a wrapper on gh to quickly get
you key Github repo information you need.The code here is used within
Roche to quickly let me pull answer simple questions like:
- How many studies have more than 1 data scientist (and roughly what’s the commit split)
- What are the common languages being used (proxied through file type distribution within repos)
- Pull commit metadata to enrich other study info held in other systems
Table of Contents
Setup
# devtools::install_github("OpenPharma/GithubMetrics")
library(GithubMetrics)
library(glue)
library(tidyverse)
organisation <- "openpharma"
Info on the repos
Quickly pull info on all the repos in a particular org. Here I look at the organisation called OpenPharma.
repos <- organisation %>%
gh_repos_get() %>%
gh_repos_clean()
repos %>%
mutate(days_since_updated = Sys.Date() - as.Date(updated_at)) %>%
arrange(days_since_updated) %>% select(name,language,MB,days_since_updated) %>%
knitr::kable()
| name | language | MB | days_since_updated |
|---|---|---|---|
| GithubMetrics | R | 0.1 | 0 days |
| BBS-causality-training | R | 0.0 | 1 days |
| visR | HTML | 20.8 | 1 days |
| facetsr | R | 2.1 | 61 days |
| CTP | R | 0.9 | 85 days |
| simaerep | R | 77.6 | 86 days |
| ReadStat | C | 1.8 | 126 days |
| visR-docs | Unsure | 5.3 | 131 days |
| sas7bdat | Python | 0.1 | 141 days |
| syntrial | R | 0.3 | 199 days |
| icd_hierarchies | Unsure | 0.0 | 267 days |
| pypharma_nlp | Jupyter Notebook | 28.0 | 289 days |
| RDO | R | 0.5 | 327 days |
| openpharma.github.io | JavaScript | 0.9 | 1315 days |
Get all commits
Now I can pull all the commits on the main branch across repos in that org.
repo_all_commits <- repos %>%
filter(size > 0) %>% # make sure has some commits
pull(full_name) %>%
gh_commits_get(
days_back = 365*10
)
## Pulling commits looking back to 2011-02-02
repo_all_commits %>%
filter(!author %in% c(".gitconfig missing email","actions-user")) %>%
mutate(
repo = gsub("openpharma/","",full_name)
) %>%
group_by(repo) %>%
summarise(
commits = n(),
contributors = n_distinct(author),
last_commit = max(as.Date(datetime))
) %>% arrange(desc(commits)) %>%
knitr::kable()
| repo | commits | contributors | last_commit |
|---|---|---|---|
| ReadStat | 992 | 13 | 2020-09-04 |
| visR | 380 | 14 | 2020-11-19 |
| pypharma_nlp | 110 | 1 | 2020-04-16 |
| sas7bdat | 86 | 7 | 2020-09-10 |
| RDO | 42 | 1 | 2020-03-09 |
| GithubMetrics | 19 | 1 | 2021-01-30 |
| CTP | 6 | 3 | 2020-10-19 |
| simaerep | 6 | 1 | 2020-11-05 |
| BBS-causality-training | 3 | 1 | 2021-01-29 |
| visR-docs | 3 | 1 | 2020-09-21 |
| openpharma.github.io | 2 | 1 | 2017-06-25 |
| syntrial | 2 | 1 | 2020-07-15 |
| facetsr | 1 | 1 | 2020-11-30 |
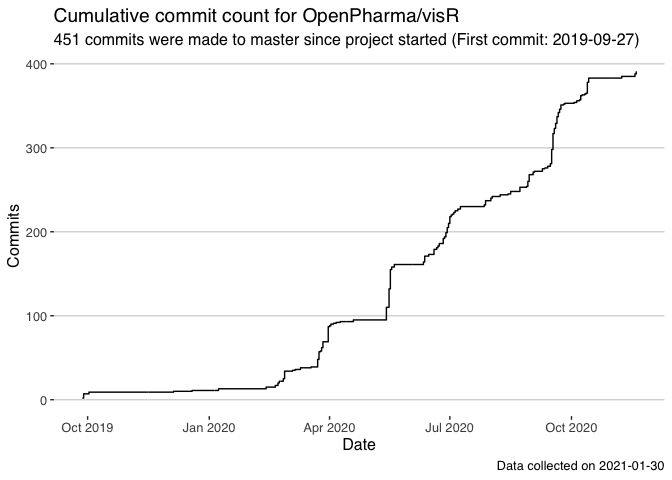
Get visR commits
Now digging into a single repo, for the R package visR.
visr_all_commits <- "OpenPharma/visR" %>%
gh_commits_get(
days_back = 365*10
) %>%
mutate(date = as.Date(datetime))
## Pulling commits looking back to 2011-02-02
visr_all_commits %>%
filter(!author %in% c(".gitconfig missing email")) %>%
ggplot(aes(x = date)) +
stat_bin(aes(y=cumsum(..count..)),geom="step", binwidth = 1) +
ggthemes::theme_hc() +
labs(
x = "Date",
y = "Commits",
title = "Cumulative commit count for OpenPharma/visR",
subtitle =
glue("{nrow(visr_all_commits)} commits were made to master since project started (First commit: {min(visr_all_commits$date)})"),
caption = paste0("Data collected on ",Sys.Date())
)
Who has been contributing to visR?
contributors <- visr_all_commits %>%
filter(!author %in% c(".gitconfig missing email","actions-user")) %>%
group_by(author) %>%
summarise(
commits = n()
)
contributors <- contributors %>%
left_join(
gh_user_get(contributors$author),
by = c("author"="username")
)
contributors %>%
arrange(-commits) %>%
mutate(
last_on_github = Sys.Date() - last_active,
contributor = glue('<img src="{avatar}" alt="" width="30"> {author}'),
blog = case_when(
blog == "" ~ "",
TRUE ~ as.character(glue('<a href="{blog}">link</a>'))
)
) %>%
select(contributor,commits,name,last_on_github,company,location,blog) %>%
knitr::kable(
caption = "People that have contributed to visR master"
)
| contributor | commits | name | last_on_github | company | location | blog |
|---|---|---|---|---|---|---|
| 127 | Steven Haesendonckx | 16 days | ||||
| 109 | Mark Baillie | 0 days | Basel, CH | link | ||
| 69 | James Black | 1 days | Roche | Basel, Switzerland | link | |
| 25 | 59 days | |||||
| 21 | Charlotta | 114 days | ||||
| 12 | Diego S | 251 days | ||||
| 4 | 1 days | |||||
| 3 | Tim Treis | 19 days | Heidelberg, Germany | |||
| 3 | Matt Kent | 4 days | Genesis Research | |||
| 2 | 285 days | Roche / 7N | ||||
| 2 | Thomas Neitmann | 14 days | Roche | Basel, Switzerland | link | |
| 1 | Adam Foryś | 16 days | @Roche | Warsaw, Poland | link | |
| 1 | 10 days | Remote | link | |||
| 1 | 68 days |
Explore the files present
Now use the API to explore files present in head across repos in this org. Just for fun I’ll compare R to Python files present.
repo_files <- gh_repo_files_get(
repo_commits = repo_all_commits,
only_last_commit = TRUE
)
## Pulling files in latest commit from 13 repos
repo_files %>%
group_by(repo) %>%
summarise(
Files = n(),
`R files` = sum(lang %in% "R"),
`Python files` = sum(lang %in% c("Python","Jupyter Notebook"))
) %>%
mutate(
Language = case_when(
`R files` > `Python files` ~ "R",
`R files` < `Python files` ~ "Python",
TRUE ~ "?"
)
) %>%
knitr::kable(
caption = "Types of files in the organisation"
)
| repo | Files | R files | Python files | Language |
|---|---|---|---|---|
| openpharma/BBS-causality-training | 4 | 2 | 0 | R |
| openpharma/CTP | 100 | 30 | 0 | R |
| openpharma/facetsr | 63 | 13 | 0 | R |
| openpharma/GithubMetrics | 44 | 22 | 0 | R |
| openpharma/openpharma.github.io | 76 | 1 | 0 | R |
| openpharma/pypharma_nlp | 131 | 0 | 49 | Python |
| openpharma/RDO | 105 | 11 | 0 | R |
| openpharma/ReadStat | 207 | 0 | 0 | ? |
| openpharma/sas7bdat | 8 | 0 | 2 | Python |
| openpharma/simaerep | 145 | 32 | 0 | R |
| openpharma/syntrial | 67 | 24 | 0 | R |
| openpharma/visR | 177 | 81 | 0 | R |
| openpharma/visR-docs | 185 | 0 | 0 | ? |
Search for code
And as a toy example of searching for code. Note that it is a plain text search, so there will be false positives, particularly if the package name is common (I think here that’s less of an issue).
helper_gh_repo_search <- function(x, org = "openpharma"){
## Slow it down! as search has 30 calls a minute rate limit.
## If you prem the search rate limit is higher, so usually not needed
if(interactive()){message("Wait 5 seconds")}
Sys.sleep(5)
## End slow down
results <- gh_repo_search(
code = x,
organisation = org
)
if(is.na(results)) {
results <- return()
}
results %>%
mutate(Package = x, Organisation = org) %>%
group_by(Organisation,Package) %>%
summarise(
Repos = n_distinct(full_name), .groups = "drop"
)
}
packages <- c(
"tidyverse","pkgdown","dplyr","data.table"
)
package_use <- bind_rows(
packages %>%
map_df(
helper_gh_repo_search, org = "openpharma"
),
packages %>%
map_df(
helper_gh_repo_search, org = "AstraZeneca"
),
packages %>%
map_df(
helper_gh_repo_search, org = "Roche"
),
packages %>%
map_df(
helper_gh_repo_search, org = "Genentech"
),
packages %>%
map_df(
helper_gh_repo_search, org = "Novartis"
)
)
## tidyverse does not appear in AstraZeneca.
## pkgdown does not appear in AstraZeneca.
## data.table does not appear in AstraZeneca.
## query = 'data.table in:file user:AstraZeneca'
package_use %>%
pivot_wider(names_from = "Package", values_from = "Repos") %>%
mutate(Total = rowSums(.[,-1], na.rm = TRUE)) %>%
arrange(-Total) %>%
knitr::kable(
caption = "Package use detected within repositaries in Pharma orgs"
)
| Organisation | tidyverse | pkgdown | dplyr | data.table | Total |
|---|---|---|---|---|---|
| Novartis | 4 | 6 | 10 | 12 | 32 |
| openpharma | 4 | 6 | 6 | 2 | 18 |
| Roche | 3 | 3 | 2 | 3 | 11 |
| Genentech | 3 | 2 | 3 | 3 | 11 |
| AstraZeneca | 1 | 1 |